A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction
Author
Victor Iapascurta
Title
A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction
Description
The code for data processing
Category
Academic Articles & Supplements
Keywords
signal processing, block decomposition method, sepsis, machine learning
URL
http://www.notebookarchive.org/2022-03-b3yfik0/
DOI
https://notebookarchive.org/2022-03-b3yfik0
Date Added
2022-03-24
Date Last Modified
2022-03-24
File Size
11.85 megabytes
Supplements
Rights
Redistribution rights reserved

Victor Iapăscurtă, “A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction.” In: Tiginyanu, I., Sontea, V., Railean, S. (eds) 5th International Conference on Nanotechnologies and Biomedical Engineering. ICNBME 2021. IFMBE Proceedings, vol 87. Springer, Cham. https://doi.org/10.1007/978-3-030-92328-0_29.
A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction
A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction
Victor Iapăscurtă
Department of Anesthesia and Intensive Care, N. Testemițanu State University of Medicine and Pharmacy, Chișinău, Moldova
Most of the data generated by monitors in a clinical setting represent time series data which can be visualized and subsequently used for decision making . This usually is the simplest part . A more challenging aspect is using this data for more complex task like machine learning with the same goal – computer assisted decisions . Within this challenge raw biomedical signal data need to be preprocessed before being passed to the machine learning algorithm . This can be done by a multitude of methods . A number of such methods comes from the field of Algorithmic Complexity and although of a promising nature, these particular methods are poorly explored yet . The current research presents an example of applying the Block Decomposition Method to data routinely generated by patients in a modern Intensive Care Unit . The final goal of a larger research, the actual research being part of, is building a system for early sepsis prediction .
The data set used in the research represents public access data from the ' Early Prediction of Sepsis from Clinical Data - the PhysioNet Computing in Cardiology Challenge 2019' (https://physionet.org/content/challenge -2019/1.0.0) .
Final set of the data to used for Machine Learning (ML) is based on six physiological parameters : heart rate (HR), oxygen saturation (SaO2), systolic blood pressure (SBP), diastolic blood pressure (DBP), respiratory rate (RR) and temperature (Temp) . Before being fed to an ML algoritm the Kolmogorov - Chaitin complexity of data is estimated (on 3 x 3 matrices) with the use of a Complexity Calculator based on the Block Decomposition Method (https : // www . algorithmicdynamics . net/software . html) .
A final data sample for ML represents a 14 L vector where the fist two elements denote the complexity of two matrices (based on the six parameters values for three hourly observations), twelve elements with the difference of respective parameters value between 2 consecutive hours . Every samplee is labeled (sepsis - 1, non - sepsis - 0) .
Final set of the data to used for Machine Learning (ML) is based on six physiological parameters : heart rate (HR), oxygen saturation (SaO2), systolic blood pressure (SBP), diastolic blood pressure (DBP), respiratory rate (RR) and temperature (Temp) . Before being fed to an ML algoritm the Kolmogorov - Chaitin complexity of data is estimated (on 3 x 3 matrices) with the use of a Complexity Calculator based on the Block Decomposition Method (https : // www . algorithmicdynamics . net/software . html) .
A final data sample for ML represents a 14 L vector where the fist two elements denote the complexity of two matrices (based on the six parameters values for three hourly observations), twelve elements with the difference of respective parameters value between 2 consecutive hours . Every samplee is labeled (sepsis - 1, non - sepsis - 0) .
Loading preprocessed data :
In[]:=
dataImport=Import["https://docs.google.com/spreadsheets/d/e/2PACX-1vTTVcVBMLyRTvCkjnXq3ytUl0G2VHox7WG0VwcQyAkavDtP1ontENhLs7Z4evihAHSnuHzsRD4hPTem/pub?gid=41776014&single=true&output=csv"]
Out[]=
{{15.7874,16.3618,-3,0,0.02,4,-1,-0.36,-5,0,0.03,-2,2,-0.35,1},{17.6237,16.8128,7,0,0.39,-2.3,-3.7,1,4,0,0.39,-2.4,-3.6,0.5,1}, ⋯31833⋯ | |||||
|
Note : In this notebook an updated/larger data set that contains 31837 samples (vs the set in the article) is used .
In[]:=
n=Length[dataImport]
Out[]=
31837
Creating training and test sets:
In[]:=
trainN=Floor[0.9*n]
Out[]=
28653
In[]:=
{dataTrain,dataTest}=TakeDrop[RandomSample@dataImport,trainN];Dimensions[dataTrain]Dimensions[dataTest]
Out[]=
{28653,15}
Out[]=
{3184,15}
In[]:=
xTrain=dataTrain[[All,1;;14]];yTrain=dataTrain[[All,15]];xTest=dataTest[[All,1;;14]];yTest=dataTest[[All,15]];
In[]:=
trainingset={xTrain->yTrain};
In[]:=
testset={xTest->yTest}
Out[]=
{{{16.8532,16.2174,5.5,0,0.35,3,-1.5,0.5,1.5,-0.5,0.45,-2,-6,4.5}, ⋯3182⋯ ⋯3118⋯ | |||||
|
Machine Learning (by Gradient Boosted Trees Method) :
In[]:=
c=Classify[xTrainyTrain,Method"GradientBoostedTrees"]
Out[]=
ClassifierFunction
| |||||
Data not saved. Save now |
In[]:=
ClassifierFunction
|
Out[]=
ClassifierFunction
| |||||
Data not saved. Save now |
In[]:=
cm=ClassifierMeasurements[c,xTestyTest];
Verifying the classifier on a test sample :
In[]:=
c[{18.38286372,16.85323886,0,0,0.05,-1,1,0,-0.5,-0.5,0,27.5,8.5,-0.5},"Probabilities"]
Out[]=
00.965292,10.0347079
Displaying performance metrics :
In[]:=
plt1=cm["ConfusionMatrixPlot"];
In[]:=
Style[plt1,Magnification3]
Out[]=

In[]:=
cm["ROCCurve"]
Out[]=
0
,1
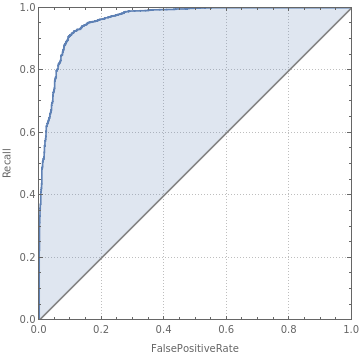 |
|
 |
|
In[]:=
cm["Accuracy"]
Out[]=
0.911432
In[]:=
cm["Sensitivity"]
Out[]=
00.944193,10.856904
In[]:=
cm["Specificity"]
Out[]=
00.856904,10.944193
In[]:=
cm["Precision"]
Out[]=
00.916545,10.902203
Cite this as: Victor Iapascurta, "A Less Traditional Approach to Biomedical Signal Processing for Sepsis Prediction" from the Notebook Archive (2022), https://notebookarchive.org/2022-03-b3yfik0
Download
