03 Compound vs Substance
Author
Joshua Schrier
Title
03 Compound vs Substance
Description
Understand the difference between compounds and substances in PubChem’s terminology.
Category
Educational Materials
Keywords
cheminformatics, Chemoinformatics, chemical information, PubChem, quantitative structure, property relationships, QSPR, machine learning, computer-aided drug design, chemistry
URL
http://www.notebookarchive.org/2020-10-ebny1gg/
DOI
https://notebookarchive.org/2020-10-ebny1gg
Date Added
2020-10-31
Date Last Modified
2020-10-31
File Size
209.28 kilobytes
Supplements
Rights
CC BY-NC-SA 4.0

Compound vs Substance
Compound vs Substance
Objectives
Objectives
◼
Understand the difference between compounds and substances in PubChem’s terminology.
◼
Learn how chemical structures are represented in a real world.
◼
Understand the ambiguities of name-structure associations.
◼
Learn how to draw chemical structures programmatically.
Structure Standardization
Structure Standardization
PubChem contains more than 200 millions chemical records submitted by hundreds of data contributors. These depositor-provided records are archived in a database called “Substance” and each record in this database is called a substance. The records in the Substance database are highly redundant, because different data contributors may submit information on the same chemical, independently of each other. Therefore, PubChem extracts unique chemical structures from the Substance database through a process called standardization (https://doi.org/10.1186/s13321-018-0293-8). These unique structures are stored in the Compound database and individuate records in this database is called “compounds”. To learn more about the PubChem compounds and substances, please read this PubChem Blog post (https://go.usa.gov/xVXct).
The code cells below demonstrates the effects of chemical structure standardization.
The code cells below demonstrates the effects of chemical structure standardization.
Step 1. Download a list of the SIDs associated with a given CID.
Step 1. Download a list of the SIDs associated with a given CID.
First, let's get a list of SIDs that are associated CID 1174 (uracil) either by manually constructing the PUG-REST call:
In[]:=
cid=1174;url=URLBuild[{"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/",ToString[cid],(*note:conversionofintegerCIDtostringtype*)"sids/txt"}]sids=URLExecute[url,{},"CSV"]//Flatten;Length[sids]
Out[]=
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1174/sids/txt
Out[]=
396
In[]:=
sids=ServiceExecute["PubChem","CompoundSID",{"CompoundID"1174}][First,"SubstanceID"]//Normal;Length[sids]
Out[]=
396
Step 2. Download the structure data for the SIDs
Step 2. Download the structure data for the SIDs
Now retrieve the depositor-provided structures for the returned substances. We'll begin by defining a function that retrieves the SDFs give a list of SID values:
retrieveSDF[sids_List]:=With[{url=URLBuild[{"https://pubchem.ncbi.nlm.nih.gov/rest/pug/substance/sid/",StringRiffle[sids,","],(*generateastringofcomma-separatedSIDs*)"/record/sdf"}]},URLExecute[url,{},"Text"]]
Next: Partition the list of SIDs into chunks of UpTo 50 elements each, and use a MapBatched operation to retrieve each of these batches, Pause-ing for 0.2 seconds between each batch. This will take a minute or two to execute:
In[]:=
chunks=Partition[sids,UpTo[50]];sdfs=
[retrieveSDF,chunks,Pause[0.2],1];
[◼] | MapBatched |
At the end, we Export a file that has joined all of the chunks together. Because the data received from PubChem is in the form of text files, we force the Export command to write this as “Text”, rather than trying to interpret the input as an SDF file. We also insert a return (“\n”) after each block of text using StringRiffle, before writing the file to disk:
In[]:=
Export["cid2sids-uracil.sdf",StringRiffle[sdfs,"\n"],"Text"]
Out[]=
cid2sids-uracil.sdf
Take a moment to open this file in a text editor to get a sense of its contents.
Step 3. Convert the structures in the SDF file into the SMILES strings and identify unique SMILES and their frequencies.
Step 3. Convert the structures in the SDF file into the SMILES strings and identify unique SMILES and their frequencies.
Begin by Import-ing the structures from the SDF file that was written to your local disk in the previous step; Import looks at the file suffix (.sdf, .sd) and interprets it as containing molecular data on Import, returning a list of Molecule representations (introduced at the end of Assignment 1):
In[]:=
mols=Import["cid2sids-uracil.sdf"];(*370+Molecule[]s!*)
Let’s look at the first of these:
In[]:=
example=First[mols]
Out[]=
Molecule
 |
|
In[]:=
MoleculeProperty["SMILES"]@example
Out[]=
c1c[nH]c(=O)[nH]c1=O
These properties also include any metadata contained in the SDF entry:
In[]:=
MoleculeProperty["MetaInformation"]@example
Out[]=
PUBCHEM_CID_ASSOCIATIONS{1174,1},PUBCHEM_COMPOUND_ID_TYPE0,PUBCHEM_COORDINATE_TYPE{1,3},PUBCHEM_EXT_DATASOURCE_NAMEBioCyc,PUBCHEM_EXT_DATASOURCE_REGIDURACIL,PUBCHEM_EXT_DATASOURCE_URLhttps://biocyc.org/,PUBCHEM_EXT_SUBSTANCE_URLhttps://biocyc.org/compound?orgid=META&id=URACIL,PUBCHEM_GENERIC_REGISTRY_NAME66-22-8,PUBCHEM_SUBSTANCE_COMMENT{TYPES: Pyrimidine-Bases, Pyrimidines,Is a product of enzyme EC 3.2.2.27,Is a product of enzyme EC 3.2.2.28,Is a product of enzyme EC 2.4.2.9,Is a product of enzyme EC 2.4.2.2,Is a product of enzyme EC 3.5.4.1,Is a product of enzyme EC 2.4.2.3,Is a reactant of enzyme EC 4.2.1.70,Is a reactant of enzyme EC 1.17.99.4,Is a product of enzyme EC 1.14.11.10,Is a reactant of enzyme EC 1.14.99.46,Is a product of enzyme EC 1.3.3.7,Is a reactant of enzyme EC 2.5.1.53,Is a product of enzyme EC 3.13.1.M4,Is a product of enzyme EC 2.4.2.57,Is a product of enzyme EC 3.2.2.3,Is a product of enzyme EC 1.3.1.1,Is a product of enzyme EC 4.1.1.66,Is a product of enzyme EC 1.3.1.2},PUBCHEM_SUBSTANCE_ID3272,PUBCHEM_SUBSTANCE_SYNONYM{uracil,66-22-8},PUBCHEM_SUBSTANCE_VERSION25,PUBCHEM_TOTAL_CHARGE0,PUBCHEM_XREF_EXT_IDURACIL
To identify unique SMILES and their frequencies, we’ll first obtain the SMILES property for each of the molecules in the mol list, and then obtain an Association of Counts of the number of time each string appears:
In[]:=
uniqueSmilesFreq=Counts[MoleculeProperty["SMILES"]/@mols]
Out[]=
c1c[nH]c(=O)[nH]c1=O17,O=c1[nH]c(=O)cc[nH]1169,O=c1n([H])c(=O)ccn1[H]21,O(c1nc(O[H])nc([H])c1[H])[H]32,O=c1n([H])c(=O)c([H])c([H])n1[H]30,[nH]1c(=O)[nH]c(=O)cc19,Oc1nc(O)ccn149,O=c1nc(O)cc[nH]12,O=c1[nH]ccc(=O)[nH]11,15,[nH]1c(=O)[nH]ccc1=O6,n1c(O)nccc1O2,O=c1n([H])c(=O)cc[nH]12,O=c1n([H])ccc(=O)[nH]12,[nH]1c(=O)cc[nH]c1=O4,O=c1n([H])ccc(O)n12,O=c1n([H])c(O)ccn11,Oc1n([H])c(=O)ccn11,O=c1ccn([H])c(=O)n1[H]2,Oc1nc(=O)cc[nH]13,O=c1[nH]c(O)ccn13,Oc1[nH]c(=O)ccn14,O=c1[nH]c(=O)[nH]cc11,c1(=O)[nH]c(=O)cc[nH]12,c1cnc(O)nc1O2,Oc1nccc(O)n12,c1(O)[nH]c(=O)ccn11,[nH]1c(=O)nccc1O1,n1c(O)[nH]c(=O)cc11,Oc1[nH]ccc(=O)n11,[nH]1c(=O)nc(O)cc11,n1([H])c(=O)n([H])ccc1=O1,n1([H])c(=O)n([H])c(=O)c([H])c1[H]1,n1c(O)nc(O)cc11,c1([H])c([H])nc(O[H])nc1O[H]1,[H]n1ccc(=O)n([H])c1=O1,O=c1cc[nH]c(=O)[nH]12
If you look closely at the above list, you will notice that many of these structures vary only in the order in which the atoms are included. We can remove this ambiguity (while still retaining real differences in the specified protonation state, tautomerization, etc.) by using the “CanonicalSMILES” string:
In[]:=
uniqueSmilesFreq=Counts[MoleculeProperty["CanonicalSMILES"]/@mols]
Out[]=
O=c1cc[nH]c(=O)[nH]1271,Oc1ccnc(O)n189,O=c1nc(O)cc[nH]15,15,O=c1nccc(O)[nH]15,O=c1ccnc(O)[nH]17,O=c1cc[nH]c(O)n14
In[]:=
Dataset[%]
Out[]=
|
(As noted in earlier exercises: Do not be alarmed if the numbers that you observe when you run this example are greater than the numbers shown above. New records are added to PubChem every day.)
The above output shows that the 390+ SIDs associated with CID 1174 are represented with six different SMILES strings. In addition, 15 substance records that resulted in an “empty” SMILES strings, implying that the depositors of these substance records did not provide structural information.
You may want to what these 15 substances without SMILES strings are and how the structures were generated. Begin by Select-ing the particular cases where the SMILES string is empty (because there are no atoms). In this example we will use Select in the function/argument style, where the list to be tested (“mols”) is the first argument, and the test to be applied is the pure function defined as the second argument:
You may want to what these 15 substances without SMILES strings are and how the structures were generated. Begin by Select-ing the particular cases where the SMILES string is empty (because there are no atoms). In this example we will use Select in the function/argument style, where the list to be tested (“mols”) is the first argument, and the test to be applied is the pure function defined as the second argument:
In[]:=
noStructure=Select[mols,StringMatchQ[""]@MoleculeProperty["SMILES"]@#&];
Then define a function that takes a single Molecule as input and returns the information of interest as a list:
In[]:=
reportMissing[mol_Molecule]:=With[{metadata=MoleculeProperty["MetaInformation"]@mol},{metadata["PUBCHEM_SUBSTANCE_ID"],metadata["PUBCHEM_SUBS_AUTO_STRUCTURE"]}]
(a complete list of possible information available is documented at ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_sdtags.txt )
Finally, Map this function over the list of structure-less molecules determined above, and display the result in TableForm for aesthetic purposes:
Finally, Map this function over the list of structure-less molecules determined above, and display the result in TableForm for aesthetic purposes:
In[]:=
TableForm[reportMissing/@noStructure]
Out[]//TableForm=
50608295 | Deposited Substance chemical structure was generated via Synonym "CID1174" to be CID 1174 |
76715622 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
313082517 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
319449334 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
329735657 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
330000149 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
376019281 | Deposited Substance chemical structure was generated via Synonym(s) "2,4(1H,3H)-PYRIMIDINEDIONE", "66-22-8" and Synonym Consistency to be CID 1174 |
381002398 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
381013941 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
381360788 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
384257697 | Deposited Substance chemical structure was generated via Synonym(s) "uracil" and MeSH to be CID 1174 |
384995482 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
402318513 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
402318514 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
402318515 | Deposited Substance chemical structure was generated via Synonym(s) "66-22-8" and Synonym Consistency to be CID 1174 |
Step 4. Generate the structure images from the SMILES
Step 4. Generate the structure images from the SMILES
Now we want to see what these SMILES strings look like, by drawing molecular structures from them. In the previous section, we defined an association with canonical SMILES strings as keys and the number of occurrences as the corresponding values. Let’s print it out again to recall what it looks like:
In[]:=
uniqueSmilesFreq
Out[]=
O=c1cc[nH]c(=O)[nH]1271,Oc1ccnc(O)n189,O=c1nc(O)cc[nH]15,15,O=c1nccc(O)[nH]15,O=c1ccnc(O)[nH]17,O=c1cc[nH]c(O)n14
We’ll Select from the Association the Keys whose StringLength is greater than zero (i.e., removing the cases with missing structures). In this example, we will use Select in the operator form, defining the test to be applied as its argument, and then applying it to the Keys (i.e., the SMILES strings):
In[]:=
mySmiles=Select[StringLength[#]>0&]@Keys@uniqueSmilesFreq
Out[]=
{O=c1cc[nH]c(=O)[nH]1,Oc1ccnc(O)n1,O=c1nc(O)cc[nH]1,O=c1nccc(O)[nH]1,O=c1ccnc(O)[nH]1,O=c1cc[nH]c(O)n1}
Using this list of non-blank SMILES strings, then create a new Association whose values are the corresponding 2D-structures:
In[]:=
AssociationMap[MoleculePlot@Molecule[#]&,(*functionusedtogeneratethevalues*)mySmiles(*keysused*)]
Out[]=
O=c1cc[nH]c(=O)[nH]1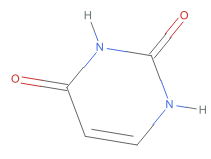 ,Oc1ccnc(O)n1
,Oc1ccnc(O)n1 ,O=c1nc(O)cc[nH]1
,O=c1nc(O)cc[nH]1 ,O=c1nccc(O)[nH]1
,O=c1nccc(O)[nH]1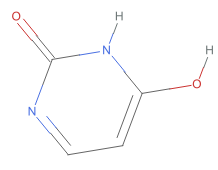 ,O=c1ccnc(O)[nH]1
,O=c1ccnc(O)[nH]1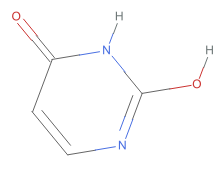 ,O=c1cc[nH]c(O)n1
,O=c1cc[nH]c(O)n1






You may want to write these molecule images in files, rather than displaying them on this Mathematica notebook:
In[]:=
Export["image_"<>#<>".png",MoleculePlot@Molecule[#]]&/@mySmiles
Out[]=
{image_O=c1cc[nH]c(=O)[nH]1.png,image_Oc1ccnc(O)n1.png,image_O=c1nc(O)cc[nH]1.png,image_O=c1nccc(O)[nH]1.png,image_O=c1ccnc(O)[nH]1.png,image_O=c1cc[nH]c(O)n1.png}
You may also want to display all the images in a single figure:
In[]:=
figures=MoleculePlot@Molecule[#]&/@mySmiles;GraphicsRow[figures]
Out[]=
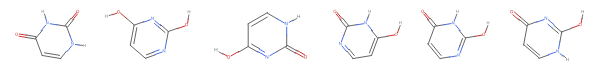
...and then save the resulting image to a file:
In[]:=
Export["image_grid.png",%]
Out[]=
image_grid.png
As shown these chemical images, the 390+ substances associated with CID 1174 (uracil) correspond to six tautomeric form of uracil, which differ from each other in the position of “movable” hydrogen atoms. Compare these structures with their standardized structure (CID 1174):
In[]:=
MoleculePlot@Molecule@URLExecute["https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1174/property/isomericsmiles/txt"]
Out[]=

Alternatively, you can get the structure image of CID 1174 directly from PubChem either by constructing the REST-PUG call yourself:
In[]:=
URLExecute["https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1174/record/PNG?image_size=300x300"]
Out[]=

Or by using Mathematica’s PubChem service wrapper to retrieve the images:
In[]:=
ServiceExecute["PubChem","CompoundImage",{"CompoundID"1174}]
Out[]=

Exercises:
Exercises:
Comment: Exercises 1a and 1b in the have been omitted because they are specific to RDKit.
Exercise 1c
Exercise 1c
Retrieve the substance records associated with guanine (CID 135398634) and display unique structures generated from them, by following these steps:
◼
Retrieve the SIDs associated CID 135398634
◼
Download the structure data for the retrieved SIDs (in SDF).
◼
Generate canonical SMILES strings from the structure data in the SDF file and identify unique canonical SMILES strings.
◼
Draw the structures represented by the unique canonical SMILES strings in a single figure.
In[]:=
(*Writeyourcodeinthiscell.*)
Exercise 1d
Exercise 1d
Retrieve the substance records whose synonym is “glucose” and display unique structures generated from them, by following these steps:
◼
Retrieve the SIDs whose synonym is “glucose”.
◼
Download the structure data for the retrieved SIDs (in SDF).
◼
Generate canonical SMILES strings from the structure data in the SDF file and identify unique canonical SMILES strings
◼
Draw the structures represented by the unique canonical SMILES strings in a single figure.
In[]:=
(*Writeyourcodeinthiscell.*)
Exercise 1e
Exercise 1e
Retrieve the compound records associated with the SIDs retrieved in Exercise 1d and display unique structures generated from them, by following these steps:
◼
Retrieve the CIDs associated with the SIDs whose name is “glucose”, using a single PUG-REST request (as discussed in Assignment 2).
◼
◼
Retrieve the isomeric SMILES for the unique CIDs through PUG-REST
◼
Draw the structures represented by the returned SMILES strings in a single figure.
In[]:=
(*Writeyourcodeinthiscell.*)
Attributions
Attributions
Adapted from the corresponding OLCC 2019 Python Assignment:
https://chem.libretexts.org/Courses/Intercollegiate_Courses/Cheminformatics_OLCC_ (2019)/3._Database _Resources _in _Cheminformatics/3.7%3 A_Python _Assignment
https://chem.libretexts.org/Courses/Intercollegiate_Courses/Cheminformatics_OLCC_ (2019)/3._Database _Resources _in _Cheminformatics/3.7%3 A_Python _Assignment
Cite this as: Joshua Schrier, "03 Compound vs Substance" from the Notebook Archive (2020), https://notebookarchive.org/2020-10-ebny1gg
Download
